Paper Review: the Babylon Chatbot
June 29, 2018 § 2 Comments
[For convenience I collect here and slightly rearrange and update my Twitter review of a recent paper comparing the performance of the Babylon chatbot against human doctors. As ever my purpose is to focus on the scientific quality of the paper, identify design weaknesses, and suggest improvements in future studies.]
Here is my peer review of the Babylon chatbot as described in the conference paper at https://marketing-assets.babylonhealth.com/press/BabylonJune2018Paper_Version1.4.2.pdf
Please feel free to correct any misunderstandings I have of the evaluation in the tweets that follow.
To begin, the Babylon engine is a Bayesian reasoner. That’s cool. Not sure if it qualifies as AI.
The evaluation uses artificial patient vignettes which are presented in a structured format to human GPs or a Babylon operator. So the encounter is not naturalistic. It doesn’t test Babylon in front of real patients.
In the vignettes, patients were played by GPs, some of whom were employed by Babylon. So they might know how Babylon liked information to be presented and unintentionally advantaged it. Using independent actors, or ideally real patients, would have had more ecological validity.
A human is part of the Babylon intervention because a human has to translate the presented vignette and enter it into Babylon. In other words, the human heard information that they then translated into terminology Babylon recognises. The impact of this human is not explicitly measured. For example, rather than being a ‘mere’ translator, the human may occasionaly have had to make clinical inferences to match case data to Babylon capabilty. The knowledge to do that would thus be be external to Babylon and yet contribute to its performance, if true.
The vignettes were designed to test know capabilities of the system. Independently created vignettes exploring other diagnoses would likely have resulted in a much poorer performance. This tests Babylon on what it knows not what it might find ‘in the wild’.
It seems the presentation of information was in the OSCE format, which is artificial and not how patients might present. So there was no real testing of consultation and listening skills that would be needed to manage a real world patient presentation.
Babylon is a Bayesian reasoner but no information was presented on the ‘tuning’ of priors required to get this result. This makes replication hard. A better paper would provide the diagnostic models to allow independent validation.
The quality of differential diagnoses by humans and Babylon was assessed by one independent individual. In addition two Babylon employees also rated differential diagnosis quality. Good research practice is to use multiple independent assessors and measure inter-rater reliability.
The safety assessment has the same flaw. Only 1 independent assessor was used and no inter-rater reliability measures are presented when several in-house assessors are added. Non-independent assessors bring a risk of bias.
To give some further evaluation, additional vignettes are used, based on MRCGP tests. However any vignettes outside of the Babylon system’s capability were excluded. They only tested Babylon on vignettes it had a chance to get right.
So, whilst it might be ok to allow Babylon to only answer questions that it is good at for limited testing, the humans did not have a reciprocal right to exclude vignettes they were not good at. This is fundamental bias in the evaluation design.
A better evaluation model would have been to draw a random subset of cases and present them to both GPs and Babylon.
No statistical testing is done to check if the differences reported are likely due to chance variation. A statistically rigorous study would estimate the likely effect size and use that to determine the sample size needed to detect a difference between machine and human.
For the first evaluation study, methods tells us “The study was conducted in four rounds over consecutive days. In each round, there were up to four “patients” and four doctors.” That should mean each doctor and Babylon should have seen “up to” 16 cases.
Table 1 shows Babylon used on 100 vignettes and doctors typically saw about 50. This makes no sense. Possibly they lump in the 30 Semigran cases reported separately but that still does not add up. Further as the methods for Semigran were different they cannot be added in any case.
There is a problem with Doctor B who completes 78 vignettes. The others do about 50. Further looking at Table 1 and Fig 1 Doctor B is an outlier, performing far worse than the others diagnostically. This unbalanced design may mean average doctor performance is penalised by Doctor B or the additional cases they saw.
Good research practice is to report who study subjects are and how they were recruited. All we know is that these were locum GPs paid to do the study. We should be told their age, experience and level of training, perhaps where they were trained, whether they were independent or had a prior link to the researchers doing the study etc. We would like to understand if B was somehow “different” because their performance certainly was.
Removing B from the data set and recalculating results shows humans beating Babylon on every measure in Table 1.
With such a small sample size of doctors, the results are thus very sensitive to each individual case and doctor, and adding or removing a doctor can change the outcomes substantially. That is why we need statistical testing.
There is also a Babylon problem. It sees on average about twice as many cases as the doctors. As we have no rule provided for how the additional cases seen by Babylon were selected, there is a risk of selection bias eg what if by chance the ‘easy’ cases were only seen by Babylon?
A better study design would allocate each subject exactly the same cases, to allow meaningful and direct performance comparison. To avoid known biases associated with presentation order of cases, case allocation should be random for each subject.
For the MRCGP questions Babylon’s diagnostic accuracy is measured by its ability to identify a disease within its top 3 differential diagnoses. It identified the right diagnosis in its top 3 in 75% of 36 MRCGP CSA vignettes, and 87% of 15 AKT vignettes.
For the MRCGP questions we are not given Babylon’s performance when the measure is the top differential. Media reports compare Babylon against historical MRCGP human results. One assumes humans had to produce the correct diagnosis, and were not asked for a top 3 differential.
There is huge significance clinically in putting a disease in your top few differential diagnoses and the top one you elect to investigate. It also is an unfair comparison if Babylon is rated by a top 3 differential and humans by a top 1. Clarity on this aspect would be valuable.
In closing, we are only ever given one clear head to head comparison between humans and Babylon, and that is on the 30 Semigran cases. Humans outperform Babylon when the measure is the top diagnosis. Even here though, there is no statistical testing.
So, in summary, this is a very preliminary and artificial test of a Bayesian reasoner on cases for which it has already been trained.
In machine learning this would be roughly equivalent to in-sample reporting of performance on the data used to develop the algorithm. Good practice is to report out of sample performance on previously unseen cases.
The results are confounded by artificial conditions and use of few and non-independent assessors.
There is lack of clarity in the way data are analysed and there are numerous risks of bias.
Critically, no statistical testing is performed to tell us whether any of the differences seen mean anything. Further, the small sample size of GPs tested likely means it would be unlikely that this study was adequately powered to see any difference, if it does exist.
So, it is fantastic that Babylon has undertaken this evaluation, and has sought to present it in public via this conference paper. They are to be applauded for that. One of the benefits of going public is that we can now provide feedback on the study’s strength and weaknesses.
Journal Review: Watson for Oncology in Breast Cancer
March 9, 2018 § Leave a comment
How should we interpret research reporting the performance of #AI in clinical practice?
[This blog collects together in one place a twitter review published 9 March 2018 at https://twitter.com/EnricoCoiera/status/971886744101515265 ]
Today we are reading “Watson for Oncology and breast cancer treatment recommendations: agreement with an expert multidisciplinary tumor board” that has just appeared in the Annals of Oncology.
https://academic.oup.com/annonc/article/29/2/418/4781689
This paper studies the degree of agreement or “concordance” between Watson for Oncology (WFO) and an expert panel of clinicians on a ’tumor board’. It reports an impressive 93% concordance between human experts and WFO when recommending treatment for breast cancer.
Unfortunately the paper is not open access but you can read the abstract. I’d suggest reading the paper before you read further into this thread. My question to you: Do the paper’s methods allow us to have confidence in the impressive headline result?
We should begin by congratulating the authors on completing a substantial piece of work in an important area. What follows is the ‘review’ I would have written if the journal had asked me. It is not a critique of individuals or technology and it is presented for educational purposes.
Should be believe the results are valid, and secondly do we believe that they generalizable to other places or systems? To answer this we need to examine the quality of the study, the quality of the data analysis, and the accuracy of the conclusions drawn from the analysis.
I like to tease apart the research methods section using PICO headings – Population, Intervention, Comparator, Outcome.
(P)opulation. 638 breast cancer patients presented between 2014-16 at a single institution. However the study excluded patients with colloid, adenocystic, tubular, or secretory breast cancer “since WFO was not trained to offer treatment recommendations for these tumor types”.
So we have our first issue. We are not told how representative this population is of the expected distribution of breast cancer cases at the hospital or in the general population. We need to know if these 3 study years were somehow skewed by abnormal presentations.
We also need to know if this hospital’s case-mix is normal or somehow different to that in others. Its critical because any claim for the results here to generalize elsewhere depends on how representative the population is.
Also, what do we think of the phrase “since WFO was not trained to offer treatment recommendations for these tumor types”? It means that irrespective of how good the research methods are, the result will not necessarily hold for *all breast cancer cases”
All they can claim is that any result holds for this subset of breast cancers . We have no evidence presented to suggest that performance would be the same on the excluded cancers. Unfortunately the abstract and paper results section do not include this very important caveat.
(I)ntervention. I’m looking for a clear description of the intervention to understand 1/ Could someone replicate this intervention independently to validate the results? 2/ What exactly was done so that we can connect cause (intervention) with effect (the outcomes)?
WFO is the intervention. But it is never explicitly described. For any digital intervention, even if you don’t tell me what is inside the black box, I need 2 things: 1/ Describe exactly the INPUT into the system and 2/ describe exactly the OUTPUT from the system.
This paper unfortunately does neither. There is no example of how a cancer case is encoded for WFO to interpret, nor is there is an example of a WFO recommendation that humans need to read.
So the intervention is not reported in enough detail for independent replication, we do not have enough detail to understand the causal mechanism tested, and we don’t know if biases are hidden in the intervention. This makes it hard to judge study validity or generalizability
Digging into an online appendix, we do discover some details about input, and WFO mechanism. It appears WFO is input a feature vector:

It also appears that vector is then input to an unspecified machine learning engine that presumably associates disease vectors with treatments.

So, for all the discussion of WFO as a text-processing machine, at its heart might be a statistical classification engine. It’s a pity we don’t know any more, and it’s a pity we don’t know how much human labour is hidden in preparing feature vectors and training cases.
But there is enough detail in the online appendix to get a feeling about what was in general done. They really should have been much more explicit and included the input feature vector, and output treatment recommendation description in the main paper.
But was WFO really the only intervention? No. there was also a human intervention that needs to be accounted for and it might very well have been responsible for some % of the results.
The Methods reports that two humans (trained senior oncology fellows) entered the data manually into WFO. They read the cases, identified the data that matched the input features, and then decided the scores for each feature. What does this mean?
Firstly, there was no testing of inter-rater reliability. We don’t know if the two humans coded the same cases in the same way. Normally a kappa statistic is provided to measure the degree of agreement between humans and allow us to get a sense for how replicable what they did was.
For example, a low kappa means that agreement is low and that therefore any results are less likely to replicate in a future study. We really need that kappa to trust the methods were robust.
If humans are pre-digesting data for WFO how much of WFO’s performance is due to the way humans seek, identify and synthesize data? We don’t know. One might argue that the hard information detection and analysis task is done by humans and the easier classification task by WFO.
So far we have got to the point where the papers’ results are not that WFO had a 93% concordance with human experts, but rather that, when humans from a single institution read cancer cases from that institution, and extract data specifically in the way that WFO needs it, and also when a certain group of breast cancers are excluded, then concordance is 93%. That is quite a list of caveats already.
(C)ompartor: The treatments recommended by WFO are compared to the consensus recommendations of a human group of experts. The authors rightly noted that treatments might have changed between the time the humans recommended a treatment and WFO gave its recommendation. So they calculate concordance *twice*.
The first comparison is between treatment recommendations of the tumor board and WFO. The second is not so straightforward. All the cases in the first comparison for which there was no human/WFO agreement were taken back to the humans, who were asked if their opinion had changed since they last considered the case. A new comparison was then made between this subset of cases and WFO, and the 93% figure comes from this 2nd comparison.
We now have a new problem. You know you are at risk of introducing bias in a study if you do something to one group that is different to what you do to the other groups, but still pool the results. In this case, the tumor board was asked to re-consider some cases but not others.
The reason for not looking at cases for which there had been original agreement could only be that we can safely assume that the tumor board’s views would not have changed over time. The problem is that we cannot safely assume that. There is every reason to believe that for some of these cases, the board would have changed its view.
As a result, the only outcomes possible at the second comparisons are either no change in concordance or improvement in concordance. The system is inadvertently ’rigged’ to prevent us discovering there was a decrease in concordance over time because the cases that might show a decrease are excluded from measurement.
From my perspective as a reviewer that means I can’t trust the data in the second comparison because of a high risk of bias. The experiment would need to be re-run allowing all cases to be reconsidered. So, if we look at the WFO concordance rate at first comparison, which is now all I think we reasonably can look at, it drops from 93% to 73% (Table 2).
(O)utcome: The outcome of concordance is a process not a clinical outcome. That means that it measures a step in a pathway, but tells us nothing about what might have happened to a patient at the end of the pathway. To do that we would need some sort of conversion rate.
For example if we knew for x% of cases in which WFO suggested something humans had not considered, that humans would change their mind, this would allow us to gauge how important concordance was in shaping human decision making. Ideally we would like to know a ‘number needed to treat’ i.e. how many patients need to have their case considered by WFO for 1 patient to materially benefit e.g. live rather than die.
So whilst process outcomes are great early stepping-stones in assessing clinical interventions, they really cannot tell us much about eventual real world impact. At best they are a technical checkpoint as we gather evidence that a major clinical trial is worth doing.
In this paper, concordance is defined as a tricky composite variable. Concordance = all those cases for which WFO’s *recommendation* agreed with the human recommendation + all those cases in which the human recommendation appeared in a secondary WFO list of *for consideration* treatments.
The very first question I now want to know is ‘how often did human and WFO actually AGREE on a single treatment?” Data from the first human-WFO comparison point indicates that there was agreement on the *recommended* treatment in 46% of cases. That is a very different number to 93% or even 73%.
What is the problem with including the secondary ‘for consideration’ recommendations from WFO? In principle nothing, as we are at this point only measuring process not outcomes, but it might need to be measured in a very different way than it is at present for the true performance to be clear.
Our problem is that if we match a human recommended treatment to a WFO recommendation it is a 1:1 comparison. If we match human recommended to WFO ‘for consideration’ it is 1:x. Indeed from the paper I don’t know what x is. How long is this additional list of ‘for consideration’ treatments. 2,5,10? You can see the problem. If WFO just listed out the next 10 most likely treatments, there is a good chance the human recommended treatment might appear somewhere in the list. If it had listed just 2 that would be more impressive.
In information retrieval, there are metrics that deal with this type of data, and they might have been used here. You could for example report the median rank at which WFO ‘for considerations’ matched human recommendations. IN other words, rather than treating it as a binary yes/no we would better assess performance by measuring where in the list the correct recommendation is made.
Now lets us turn to the results proper. I have already explained my concerns about the headline results around concordance.
Interestingly concordance is reportedly higher for stage II and III disease and lower for stage 1 and 1V. That is interesting, but odd to me. Why would this be the case? Well, this study was not designed to test for whether concordance changes with stage – this is a post-hoc analysis. To do such a study, we would likely have had to have seek equal samples of each stage of disease. Looking at the data, stage I cases are significantly underrepresented compared to other stages. So interesting, but treat with caution.
There are other post-hoc analyses of receptor status and age and the relationship with concordance. Again this is interesting but post-hoc so treat it with caution.
Interestingly concordance decreased with age – and that might have something to do with external factors like co-morbidity starting to affect treatment recommendations. Humans might for example, reason that aggressive treatment may not make sense to a cancer patient with other illnesses and at a certain age. The best practice encoded in WFO might not take such preferences and nuances into account.
Limitations: The authors do a very good job overall in identifying many, but not all, of the issues I raise above. Despite these identified limitations, which I think are significant, they still see WFO’s producing a “high degree of agreement’ with humans. I don’t think the data yet supports that conclusion.
Conclusions: The authors conclude by suggest that as a result of this study WFO might be useful for centers with limited breast cancer resources. The evidence in this study doesn’t yet support such a conclusion. We have some data on concordance, but no data on how concordance affects human decisions, and no data on how changed decisions affects patient outcomes. Those two giant evidence gaps mean it might be a while before we safely trial WFO in real life.
So, in summary, my takeaway is that WFO generated the same treatment recommendation as humans for a subset of breast cancers at a single institution in 46% of cases. I am unclear how much influence human input had in presenting data to WFO, and there is a chance the performance might have been worse without human help (e.g. if WFO did its own text processing).
I look forward to hearing more about WFO and similar systems in other studies, and hope this review can help in framing future study designs. I welcome comments on this review.
[Note: There is rich commentary associated with the original Twitter thread, so it is worth reading if you wish to see additional issues and suggestions from the research community.]
Differences in exposure to negative news media are associated with lower levels of HPV vaccine coverage
May 1, 2017 § 1 Comment
Over the weekend, our new article in Vaccine was published. It describes how we found links between human papillomavirus (HPV) vaccine coverage in the United States and information exposure measures derived from Twitter data.
Our research demonstrates—for the first time—that states disproportionately exposed to more negative news media have lower levels of HPV vaccine coverage. What we are talking about here is the informational equivalent of: you are what you eat.
There are two nuanced things that I think make the results especially compelling. First, they show that Twitter data does a better job of explaining differences in coverage than socioeconomic indicators related to how easy it is to access HPV vaccines. Second, that the correlations are strongest for initiation (getting your first dose) than for completion (getting your third dose). If we go ahead and assume that information exposure captures something about acceptance, and that socioeconomic differences (insurance, education, poverty, etc.) signal differences…
View original post 979 more words
Making sense of consent and health records in the digital age
May 8, 2016 § 2 Comments
There are few more potent touchstones for the public than the protection of their privacy, and this is especially true with our health records. Within these documents lies information that may affect your loved ones, your social standing, employability, and the way insurance companies rate your risk.
We now live in a world where our medical records are digitised. In many nations that information is also moving away from the clinician who captured the record to regional repositories, or even government run national repositories.
The more widely accessible our records are the more likely it is that someone who needs to care for us can access them – which is good. It is also more likely that the information might be seen by individuals whom we do not know, and for purposes we would not agree with – which is the bad side of the story.
It appears that there is no easy way to balance privacy with access – any record system represents a series of compromises in design and operation that leave the privacy wishes of some unmet, and the clinical needs of others ignored.
Core to this trade-off is the choice of consent model. Patients typically need to provide their consent for their health records to be seen by others, and this legal obligation continues in the digital world.
Patient consent for others to access their digital clinical records, or e-consent, can take a number of forms. Back 2004, working with colleagues who had expertise in privacy and security, we first described the continuum of choices between patients opting in or out of consent to view their health records, as well as the trade-offs that were associated with either choice [1].
Three broad approaches to e-consent are employed.
- “Opt Out” systems; in which a population is informed that unless individuals request otherwise, their records will be made available to be shared.
- “Opt in” systems; in which patients are asked to confirm that they are happy for their records to be made available when clinicians wish to view them.
- Hybrid consent models that combine an implied consent for records to be made available and an explicit consent to view.
Opt in models assume that only those who specifically give consent will allow their health records to be visible to others, and opt out models assume that record accessibility is the default, and will only be removed if a patient actively opts-out of the process. The opt-out models maximises ease of access to, and benefit from, electronic records for clinical decision making, at the possible expense of patient privacy protections. Opt-in models have the reverse benefit, maximising consumer choice and privacy, but at the possible expense of record availability and usefulness in support of making decisions (Figure 1).
 Figure 1 – Different forms of consent balance clinical access and patient privacy in different proportions (from Coiera and Clarke, 2004)
Figure 1 – Different forms of consent balance clinical access and patient privacy in different proportions (from Coiera and Clarke, 2004)
All of the United Kingdom’s shared records systems now emply hybrid consent models of one form or another. Clinicians can also ‘break the glass’ and access records if the patient is too ill or unable to consent. In the US a variety of consent models are used and privacy legislation varies from state to state. Patients belonging to a Health Maintenance Organisation (HMO) are typically deemed to have opted in by subscribing to an HMO.
How do we evaluate the risk of one consent model over others?
The last decade has made it very clear that, at least for national systems, there are two conflicting drivers in the selection between consent models. Those that worry about patient privacy and the risks of privacy breeches favour opt-in models. Governments that worry about the political consequences of being seen to invade the privacy of their citizens thus gravitate to this model. Those that worry about having a ‘critical mass’ of consumers enrolled in their record systems, and who do not feel that they are at political risk on the privacy front (perhaps because as citizens our privacy is being so rapidly eroded on so many fronts we no longer care) seem comfortable to go the opt-out route.
The risk profiles for opt in and opt out systems are thus quite different (Figure 2). Opt-out models risk making health records available for patient’s who, in principle, would object to such access but have not opted out. This may because they were either not capable of opting-out, or were not informed of their ability to opt-out.
For opt-in models, the greatest risk to a system operator is that important clinical records are unavailable at the time of decision-making, because patients who should have elected to opt-in were neither informed that they should have a record, or were not easily capable of making that choice.
Other groups, such as those who are informed and do opt-out, may be at greater clinical risk because of that choice, but are making a decision aware of the risks.

Figure 2: The risk profiles for opt-in and opt-out patient record systems are different. Opt-out models risk making records available for patients who in principle would object to such access, but were not either capable or informed of their ability to opt-out. For opt-in models, the risk is that important clinical records are unavailable at the time of decision making, because patients who should have elected to opt-in were neither informed nor capable of making that choice.
Choosing a consent model is only half of the story
In our 2004 paper, we also made it clear that choosing between opt-in or out was not the end of the matter. There are many different ways in which we can grant access to records to clinicians and others. One can have an opt-in system which gives clinicians free access to all records with minimal auditing – a very risky approach. Alternatively you can have an opt-out system that places stringent gatekeeper demands on clinicians to prove who they are, that they have the right to access a document, that audits their access, and allows patients to specify which sections of their record are in or out – a very secure system.
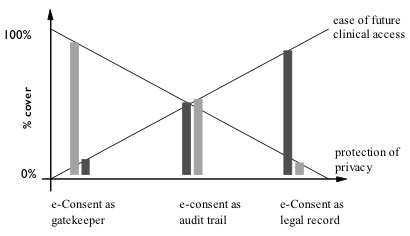
Figure 3 – The different possible functions of consent balance clinical access or patient privacy in different proportions. The diagram is illustrative of the balances only – thus there is no intention to portray the balance between access and privacy as equal in the middle model of e-Consent as an audit trail. (From Coiera and Clarke, 2004)
So, whilst we need to be clear about the risks of opt in versus opt out, we should also recognise that it is only half of the debate. It is the mechanism of governance around the consent model that counts at least as much.
For consumer advocates, “winning the war” to go opt-in is actually just the first part of the battle. Indeed, it might even be the wrong battle to be fighting. It might be even more important to ensure that there is stringent governance around record access, and that it is very clear who is reading a record, and why.
References
- Coiera E and Clarke R, e-Consent: The design and implementation of consumer consent mechanisms in an electroninc environment. J Am Med Inform Assoc, 2004. 11(2): p. 129-140.
Four futures for the healthcare system
February 20, 2016 § 1 Comment
That healthcare systems the world over are under continual pressure to adapt is not in question. With continual concerns that current arrangements are not sustainable, researchers and policy makers must somehow make plans, allocate resources, and try to refashion delivery systems as best they can.
Such decision-making is almost invariably compromised. Politics makes it hard for any form of consensus to emerge, because political consensus leads to political disadvantage for at least one of the parties. Vested interests, whether commercial or professional, also reduce the likelihood that comprehensive change will occur.
Underlying these disagreements of purpose is a disagreement about the future. Different actors all wish to will different outcomes into existence, and their disagreement means that no particular one will ever arise. The additional confounder that predicting the future is notoriously hard seems to not enter the discussion at all.
One way to minimize disagreement and to build consensus would seem to be to have all parties come to a consensus of what the future is going to be like. With a common recognition of the nature of the future that will befall us, or that we aspire to, it might becomes possible to work backwards and agree on what must happen today.
Building different scenarios to describe the future
There seem to be two major determinants of the future. The first is the environment within which the health system has to function. The second is our willingness or ability to adapt the health system to meet any particular goal or challenge. Together these two axes generate four very different future scenarios. Each scenario has a very different set of challenges to it, and very different opportunities.
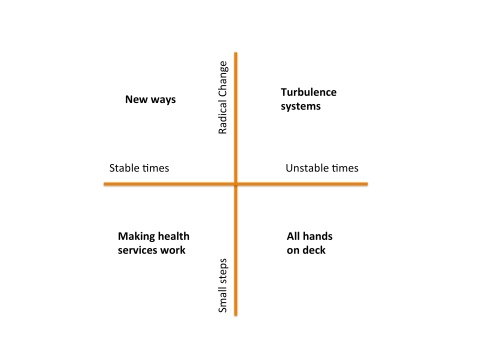
Making Health Services Work: In this quadrant, we are blessed with relatively stable conditions, and even though our capacity or will for change is modest, we can embark on incremental changes in response to projected future needs. We focus on gentle redesign of current health services, tweaking them as we need. The life of a heath services researcher is a comfortable one: no one needs or wants a revolution and there is time and resource enough to solve the problems of the day.
New Ways: Despite forgiving and stable times, in this quadrant we have an appetite for major change. Perhaps we see major changes ahead and recognize that incremental improvements will be insufficient to deal with them. Maybe we see future years with demographic challenges such as clinical workforce shortages and the increasing burden of disease associated with an ageing population. Consequently, more radical models of care are developed, evaluated and adopted. Rather than simply retro-fitting the way things are done, we radically reevaluate how things might be done, and envisage new ways of working, and conceive new ways to deliver services.
Turbulence systems: The risk of major shocks to the health system are ever present, including pandemics, weather events of ‘mass dimension’ associated with climate change, and human conflict. It is possible to make preparations for these unstable times. We might imagine that we set about to design some capacity for ‘turbulence’ management into our health services. Such turbulence systems would help us detect emerging shocks as early as possible, and would then reallocate resources as best we can when they arrive. The way that global responses to disease outbreaks has rapidly evolved over the last decade shows what is possible when our focus is on shock detection and response. Similar turbulence systems are evolving to respond to natural disasters and terrorism – so there are already models to learn from. In this quadrant then, we redesign the health system to be far more adaptive and flexible than it is today, recognizing that the future is not just going to be punctuated by rare external shocks, but that turbulence is the norm, and any system without shock absorbers will quickly shatter.
All hands on deck: In this scenario, health services receive major shocks in the near future, and well ahead of our ability to plan for these events. For example a series of major weather events or a new global pandemic could all stretch today’s health system beyond its capability to respond. Another route to this scenario in the long term is to not prepare for events like global warming or infectious disease outbreaks or an ageing population, and because of disagreement, underinvestment or poor planning, we do nothing. If such circumstances arrive, then the best thing that everyone in the health system can do is to abandon working on the long term, and apply our skills wherever they are most needed. In such crisis times, researchers will find themselves at the front lines, with a profound understanding and new respect for what implementation and translation really mean.
Picking a scenario
Which of these four worlds will we live in? It is likely that we have had the great good fortune over the last few decades of living in stable and reactively unambitious times, tinkering with a system that we have not had the appetite to change much. It seems likely that instability will increasingly become the norm however. I don’t think we will have the luxury of idly imagining some perfect but different future, debating its merits, and then starting to march toward it. There will be too much turbulence about to ever allow us the luxury of knowing exactly what the right system configuration will be. If we are very lucky, and very clever, we will increasingly redesign health services to be turbulence systems. Even if the flight to the future is a bumpy one, the stabilizers we create will help us keep the system doing what it is meant to do. ‘All hands on deck’ is the joker in the pack. I personally look forward to not ever having to work in this quadrant.
[These ideas were first published in a paper my team prepared back in 2007, and since it first appeared, the turbulence has slowly become more frequent …]
Evidence-based health informatics
February 11, 2016 § 6 Comments
Have we reached peak e-health yet?
Anyone who works in the e-health space lives in two contradictory universes.
The first universe is that of our exciting digital health future. This shiny gadget-laden paradise sees technology in harmony with the health system, which has become adaptive, personal, and effective. Diseases tumble under the onslaught of big data and miracle smart watches. Government, industry, clinicians and people off the street hold hands around the bonfire of innovation. Teeth are unfeasibly white wherever you look.
The second universe is Dickensian. It is the doomy world in which clinicians hide in shadows, forced to use clearly dysfunctional IT systems. Electronic health records take forever to use, and don’t fit clinical work practice. Health providers hide behind burning barricades when the clinicians revolt. Government bureaucrats in crisp suits dissemble in velvet-lined rooms, softly explaining the latest cost overrun, delay, or security breach. Our personal health files get passed by street urchins hand-to-hand on dirty thumbnail drives, until they end up in the clutches of Fagin like characters.
Both of these universes are real. We live in them every day. One is all upside, the other mostly down. We will have reached peak e-health the day that the downside exceeds the upside and stays there. Depending on who you are and what you read, for many clinicians, we have arrived at that point.
The laws of informatics
To understand why e-health often disappoints requires some perspective and distance. Informed observers again and again see the same pattern of large technology driven projects sucking up all the e-health oxygen and resources, and then failing to deliver. Clinicians see that the technology they can buy as a consumer is more beautiful and more useful that anything they encounter at work.
I remember a meeting I attended with Branko Cesnik. After a long presentation about a proposed new national e-health system, focusing entirely on technical standards and information architectures, Branko piped up: “Excuse me, but you’ve broken the first law of informatics”. What he meant was that the most basic premise for any clinical information system is that it exists to solve a clinical problem. If you start with the technology, and ignore the problem, you will fail.
There are many corollary informatics laws and principles. Never build a clinical system to solve a policy or administrative problem unless it is also solving a clinical problem. Technology is just one component of the socio-technical system, and building technology in isolation from that system just builds an isolated technology [3].
Breaking the laws of informatics
So, no e-health project starts in a vacuum of memory. Rarely do we need to design a system from first principles. We have many decades of experience to tell us what the right thing to do is. Many decades of what not to do sits on the shelf next to it. Next to these sits the discipline of health informatics itself. Whilst it borrows heavily from other disciplines, it has its own central reason to exist – the study of the health system, and of how to design ways of changing it for the better, supported by technology. Informatics has produced research in volume.
Yet today it would be fair to say that most people who work in the e-health space don’t know that this evidence exists, and if they know it does exist, they probably discount it. You might hear “N of 1” excuse making, which is the argument that the evidence “does not apply here because we are different” or “we will get it right where others have failed because we are smarter”. Sometimes system builders say that the only evidence that matters is their personal experience. We are engineers after all, and not scientists. What we need are tools, resources, a target and a deadline, not research.
Well, you are not different. You are building a complex intervention in a complex system, where causality is hard to understand, let alone control. While the details of your system might differ, from a complexity science perspective, each large e-health project ends up confronting the same class of nasty problem.
The results of ignoring evidence from the past are clear to see. If many of the clinical information systems I have seen were designed according to basic principles of human factors engineering, I would like to know what those principles are. If most of today’s clinical information systems are designed to minimize technology-induced harm and error, I will hold a party and retire, my life’s work done.
The basic laws of informatics exist, but they are rarely applied. Case histories are left in boxes under desks, rather than taught to practitioners. The great work of the informatics research community sits gathering digital dust in journals and conference proceedings, and does not inform much of what is built and used daily.
None of this story is new. Many other disciplines have faced identical challenges. The very name Evidence-based Medicine (EBM), for example, is a call to arms to move from anecdote and personal experience, towards research and data driven decision-making. I remember in the late ‘90s, as the EBM movement started (and it was as much a social movement as anything else), just how hard the push back was from the medical profession. The very name was an insult! EBM was devaluing the practical, rich daily experience of every doctor, who knew their patients ‘best’, and every patient was ‘different’ to those in the research trials. So, the evidence did not apply.
EBM remains a work in progress. All you need to do today is to see a map of clinical variation to understand that much of what is done remains without an evidence base to support it. Why is one kind of prosthetic hip joint used in one hospital, but a different one in another, especially given the differences in cost, hip failure and infection? Why does one developed country have high caesarian section rates when a comparable one does not? These are the result of pragmatic ‘engineering’ decisions by clinicians – to attack the solution to a clinical problem one way, and not another. I don’t think healthcare delivery is so different to informatics in that respect.
Is it time for evidence-based health informatics?
It is time we made the praxis of informatics evidence-based.
That means we should strive to see that every decision that is made about the selection, design, implementation and use of an informatics intervention is based on rigorously collected and analyzed data. We should choose the option that is most likely to succeed based on the very best evidence we have.
For this to happen, much needs to change in the way that research is conducted and communicated, and much needs to happen in the way that informatics is practiced as well:
- We will need to develop a rich understanding of the kinds of questions that informatics professionals ask every day;
- Where the evidence to answer a question exists, we need robust processes to synthesize and summarize that evidence into practitioner actionable form;
- Where the evidence does not exist and the question is important, then it is up to researchers to conduct the research that can provide the answer.
In EBM, there is a lovely notion that we need problem oriented evidence that matters (POEM) [1] (covered in some detail in Chapter 6 of The Guide to Health Informatics). It is easy enough to imagine the questions that can be answered with informatics POEMs:
- What is the safe limit to the number of medications I can show a clinician in a drop-down menu?
- I want to improve medication adherence in my Type 2 Diabetic patients. Is a text message reminder the most cost-effective solution?
- I want to reduce the time my docs spend documenting in clinic. What is the evidence that an EHR can reduce clinician documentation time?
- How gradually should I roll out the implementation of the new EHR in my hospital?
- What changes will I need to make to the workflow of my nursing staff if I implement this new medication management system?
EBM also emphasises that the answer to any question is never an absolute one based on the science, because the final decision is also shaped by patient preferences. A patient with cancer may choose a treatment that is less likely to cure them, because it is also less likely to have major side-effects, which is important given their other goals. The same obviously holds in evidence-based health informatics (EBHI).
The Challenges of EBHI
Making this vision come true would see some significant long term changes to the business of health informatics research and praxis:
- Questions: Practitioners will need develop a culture of seeking evidence to answer questions, and not simply do what they have always done, or their colleagues do. They will need to be clear about their own information needs, and to be trained to ask clear and answerable questions. There will need to be a concerted partnership between practitioners and researchers to understand what an answerable question looks like. EBM has a rich taxonomy of question types and the questions in informatics will be different, emphasizing engineering, organizational, and human factors issues amongst others. There will always be questions with no answer, and that is the time experience and judgment come to the fore. Even here though, analytic tools can help informaticians explore historical data to find the best historical evidence to support choices.
- Answers: The Cochrane Collaboration helped pioneer the development of robust processes of meta-analysis and systematic review, and the translation of these into knowledge products for clinicians. We will need to develop a new informatics knowledge translational profession that is responsible for understanding informatics questions, and finding methods to extract the most robust answers to them from the research literature and historical data. As much of this evidence does not typically come from randomised controlled trials, other methods than meta-analysis will be needed. Case libraries, which no doubt exist today, will be enhanced and shaped to support the EBHI enterprise. Because we are informaticians, we will clearly favor automated over manual ways of searching for, and summarizing, the research evidence [2]. We will also hopefully excel at developing the tools that practitioners use to frame their questions and get the answers they need. There are surely both public good and commercial drivers to support the creation of the knowledge products we need.
- Bringing implementation science to informatics: We know that informatics interventions are complex interventions in complex systems, and that the effect of these interventions vary depending on the organisational context. So, the practice of EBHI will of necessity see answers to questions being modified because of local context. I suspect that this will mean that one of the major research challenges to emerge from embracing EBHI is to develop robust and evidence-based methods to support localization or contextualisation of knowledge. While every context is no doubt unique, we should be able to draw upon the emerging lessons of implementation science to understand how to support local variation in a way that is most likely to see successful outcomes.
- Professionalization: Along with culture change would come changes to the way informatics professionals are accredited, and reaccredited. Continuing professional education is a foundation of the reaccreditation process, and provides a powerful opportunity for professionals to catch up with the major changes in science, and how those changes impact the way they should approach their work.
Conclusion
There comes a moment when surely it is time to declare that enough is enough. There is an unspoken crisis in e-health right now. The rhetoric of innovation, renewal, modernization and digitization make us all want to believers. The long and growing list of failed large-scale e-health projects, the uncomfortable silence that hangs when good people talk about the safety risks of technology, make some think that e-health is an ill-conceived if well intentioned moment in the evolution of modern health care. This does not have to be.
To avoid peak e-health we need to not just minimize the downside of what we do by avoiding mistakes. We also have to maximize the upside, and seize the transformative opportunities technology brings.
Everything I have seen in medicine’s journey to become evidence-based tells me that this will not be at all easy to accomplish, and that it will take decades. But until we do, the same mistakes will likely be rediscovered and remade.
We have the tools to create a different universe. What is needed is evidence, will, a culture of learning, and hard work. Less Dickens and dystopia. More Star Trek and utopia.
Further reading:
Since I wrote this blog a collection of important papers covering the important topic of how we evaluate health informatics and choose which technologies are fit for purpose has been published in the book Evidence-based Health Informatics.
References
- Slawson DC, Shaughnessy AF, Bennett JH. Becoming a medical information master: feeling good about not knowing everything. The Journal of Family Practice 1994;38(5):505-13
- Tsafnat G, Glasziou PP, Choong MK, et al. Systematic Review Automation Technologies. Systematic Reviews 2014;3(1):74
- Coiera E. Four rules for the reinvention of healthcare. BMJ 2004;328(7449):1197-99
An Italian translation of this article is available
A brief guide to the health informatics research literature
February 8, 2016 § Leave a comment
Every year the body of research evidence in health informatics grows. To stay on top of that research, you need to know where to look for research findings, and what the best quality sources of it are. If you are new to informatics, or don’t have research training, then you may not know where or how to look. This page is for you.
There are a large number of journals that publish only informatics research. Many mainstream health journals will also have an occasional (and important) informatics paper in them. Rather than collecting a long list of all of these possible sources, I’d like to offer the following set of resources as a ‘core’ to start with.
(There are many other very good health informatics journals, and their omission here is not meant to imply they are not also worthwhile. We just have to start somewhere. If you have suggestions for this page I really would welcome them, and I will do my best to update the list).
Texts
If you require an overview of the recent health informatics literature, especially if you are new to the area, then you really do need to sit down and read through one of the major textbooks in the area. These will outline the different areas of research, and summarise the recent state of the art.
I am of course biased and want you to read the Guide to Health Informatics.
A collection of important papers covering the important topic of how we evaluate health informatics and choose which technologies are fit for purpose can be found in the book Evidence-based Health Informatics.
Another text that has a well-earned reputation is Ted Shortliffe’s Biomedical Informatics.
Health Informatics sits on the shoulders of the information and computer sciences, psychology, sociology, management science and more. A mistake many make is to think that you can get a handle on these topics just from a health informatics text. You wont. Here are a few classic texts, from these ‘mother’ disciplines;
Computer networks (5th ed). Tannenbaum and Wetherall. Pearson. 2010.
Engineering Psychology & Human Performance (4th ed.). Wickens et al. Psychology Press. 2012.
Artificial Intelligence: A Modern Approach (3rd ed). Russell and Norvig. Pearson. 2013
Journals
Google Scholar: A major barrier to accessing the research literature is that much of it is trapped behind paywalls. Unless you work at a university and can access journals via the library, you will be asked by some publishers to pay an exorbitant fee to read even individual papers. Many journals are now however open-access, or make some of their papers available free on publication. Most journals also allow authors to freely place an early copy of a paper onto a university or other repository.
The most powerful way to finding these research articles is Google Scholar. Scholar does a great job of finding all the publicly available copies of a paper, even if the journal’s version is still behind a paywall. Getting yourself comfortable with using Scholar, and exploring what it does, provides you with a major tool for accessing the research literature.
Yearbook of Medical Informatics. The International Medical Informatics Association (IMIA) is the peak global academic body for health informatics and each year produces a summary of the ‘best’ of the last year’s research from the journals in the form of the Yearbook of Medical Informatics. The recent editions of the yearbook are all freely available online.
Next, I’d suggest the following ‘core’ journals for you to skim on a regular basis. Once you are familiar with these you will no doubt move on the the very many others that publish important informatics research.
JAMIA. The Journal of the American Medical Informatics Association (AMIA) is the peak general informatics journal, and a great place to keep tabs on recent trends. While it requires a subscription, all articles are placed into open access 12 months after publication (so you can find them using Scholar) and several articles every month are free. You can keep abreast of papers as they are published through the advanced access page.
JMIR. The Journal of Medical Internet Research is a high impact specialist journal focusing on Web-based informatics interventions. It is open access which means that all articles are free.
To round out the journals you might want to add into your regular research scan the following journals which are all very well regarded.
- Journal of Biomedical Informatics, which focuses more on methods than the other journals.
- International Journal of Medical Informatics, which tries to cover informatics issues with a global perspective.
- Artificial Intelligence in Medicine, which as the name suggest is focussed on advance topics in decision support and analytics.
- BMC Medical Informatics and Decision Making, which is an open access member of the BMC family of journals
- Methods of Information in Medicine, specialises in informatics methodologies.
Conferences
Whilst journals typically will publish well polished work, there is often a lag of a year or more before submitted papers are published. The advantage of research conferences is that you get more recent work, sometimes at an earlier stages of development, but also closer to the cutting edge.
There are a plethora of informatics conferences internationally but the following publish their papers freely online, and are typically of high quality.
AMIA Annual Symposium. AMIA holds what is probably the most prestigious annual health informatics conference, and releases all papers via NLM. An associated AMIA Summit on Translational Sciences/Bioinformatics is also freely available.
Medinfo. IMIA holds a biannual international conference, and given its status as the peak global academic society, Medinfo papers have a truly international flavour. Papers are open access and made available by IOS press through its Studies in Health Technology and Informatics series (where many other free proceedings can be found). Recent Medinfo proceedings include 2015 and 2013.
As with textbooks and journals, it is worth remembering that much of importance to health informatics is published in other ‘mother’ disciplines. For example it is well worth keeping abreast of the following conferences for recent progress:
WWW conference. The World Wide Web Conference is organised by the ACM and is an annual conference looking at innovations in the Internet space. Recent proceedings include 2015 and 2014.
The ACM Digital Library, which contains WWW, is a cornucopia of information and computer science conference proceedings. Many a rainy weekend can be wasted browsing here. You may need to hunt the web site of the actual conference however to get free access to papers as ACM will often try to charge for papers you can find freely on the home page of the conference.
Other strategies
Browsing journals is one way to keep up to date. The other is to follow the work of individual researchers whose interests mirror your own. The easiest way to do this is to find their personal page on Google Scholar (and if they don’t have one tell them to make one!). Here is mine, as an example. There are two basic ways to attack a scholar page. When you first see a Scholar page, the papers are ranked by their impact (as measured by other people citing the papers). This will give you a feeling about the work the researcher is most noted for. The second way is to click the year button. You will then see papers in date order, starting with the most recent. This is a terrific way of seeing what your pet researcher has been up to lately.
There is a regularly updated list of biomedical informatics researchers, ranked by citation impact, and this is a great way to discover health informatics scientists. Remember that when researchers work in more specialised fields, they may not have as many citations and so be lower down the list.
Once you find a few favourite researchers, try to see what they have done recently, follow them on Twitter, and if they have a blog, try to read it.